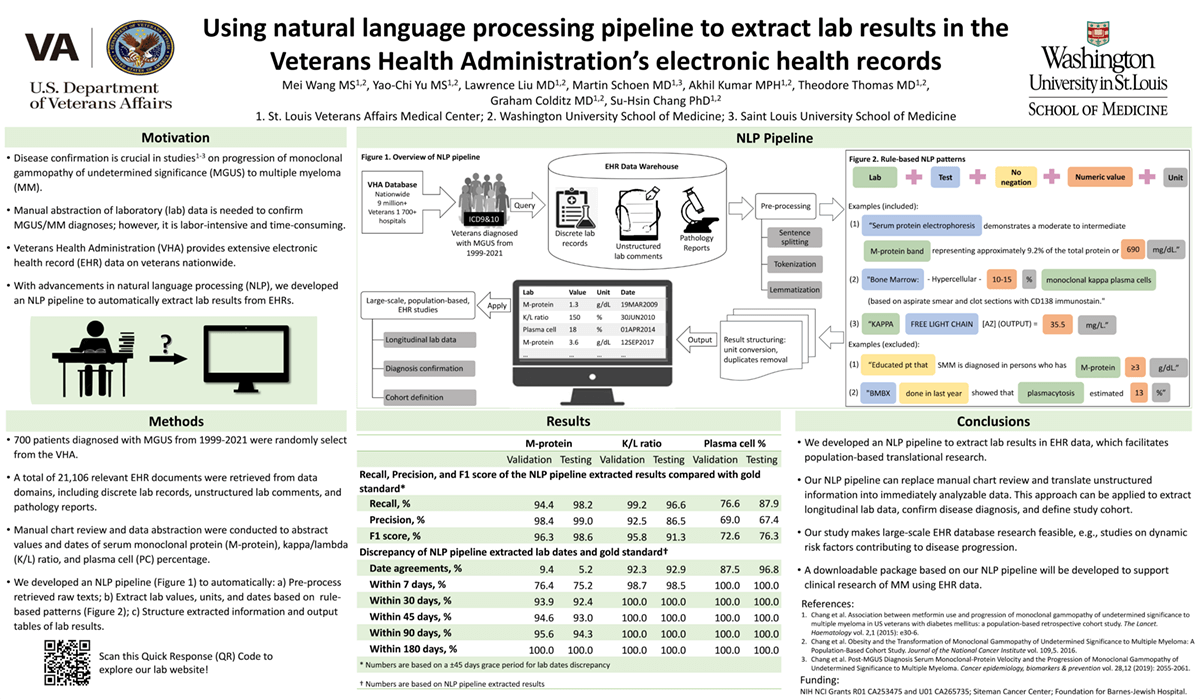
Introduction: Multiple myeloma (MM) is a common hematologic malignancy. Our studies utilized electronic health record (EHR) data in the nationwide Veteran Health Administration to study the risk of progression of monoclonal gammopathy of undetermined significance (MGUS) to MM. Disease confirmation is crucial in these studies. Relying on manual laboratory (lab) data abstraction to confirm diagnoses is labor-intensive and time-consuming, jeopardizing the feasibility of large-scale studies. With advancements in natural language processing (NLP), we developed an NLP pipeline to automate this process.
Methods: We retrieved 21,106 relevant EHR documents including discrete lab records, unstructured lab comments, and surgical pathology reports from 700 randomly selected patients diagnosed with MGUS from 1999-2021. All documents were manually reviewed to abstract the values of serum monoclonal protein (M-protein), kappa/lambda (K/L) ratio, and plasma cell (PC) % and the corresponding dates. These results were served as the reference. We then developed an NLP pipeline using pattern-based rules to extract lab values, units and dates. The performance of the NLP pipeline was compared to the reference using four metrics: recall, precision, and F1 score. The difference between NLP-extracted dates and the reference was also computed.
Results: The NLP pipeline achieved recall, precision and F1 score of 98, 99 and 99% for M-protein, 97, 87 and 91% for K/L ratio, and 88, 67 and 76% for PC %, respectively. 75, 99 and 100% of NLP-generated dates for M-protein, K/L ratio, and PC % results matched the reference within 7 days, respectively. On average, manual chart review required 15 minutes per patient to abstract all three lab results (excluding data loading time), whereas our NLP pipeline completed 20 patients per minute.
Impact: We successfully developed an NLP pipeline to extract lab results in EHR data. This approach can replace manual review and translate unstructured information into analyzable data for diagnosis confirmation. With further adaptation, our NLP pipeline may be applied to other disease areas and assist researchers in conducting large-scale EHR database research.
Organization: Washington University in St. Louis
Wang M, Yu YC, Liu L, Schoen M, Kumar A, Colditz G, Thomas T, Chang SH